Plots covariate coefficients and their confidence intervals.
# S3 method for class 'gllvm'
coefplot(
object,
y.label = TRUE,
which.Xcoef = NULL,
order = TRUE,
cex.ylab = 0.5,
cex.xlab = 1.3,
mfrow = NULL,
mar = c(4, 6, 2, 1),
xlim.list = NULL,
ind.spp = NULL,
...
)Arguments
- object
an object of class 'gllvm'.
- y.label
logical, if
TRUE(default) colnames of y with respect to coefficients are added to plot.- which.Xcoef
vector indicating which covariate coefficients will be plotted. Can be vector of covariate names or numbers. Default is
NULLwhen all covariate coefficients are plotted.- order
logical, whether or not coefficients are ordered, defaults to
TRUE.- cex.ylab
the magnification to be used for axis annotation relative to the current setting of cex.
- cex.xlab
the magnification to be used for axis annotation.
- mfrow
same as
mfrowinpar. IfNULL(default) it is determined automatically.- mar
vector of length 4, which defines the margin sizes:
c(bottom, left, top, right). Defaults toc(4,5,2,1).- xlim.list
list of vectors with length of two to define the intervals for an x axis in each covariate plot. Defaults to NULL when the interval is defined by the range of point estimates and confidence intervals
- ind.spp
vector of species indices to construct the caterpillar plot for
- ...
additional graphical arguments.
Examples
# Extract subset of the microbial data to be used as an example
data(microbialdata)
X <- microbialdata$Xenv
y <- microbialdata$Y[, order(colMeans(microbialdata$Y > 0),
decreasing = TRUE)[21:40]]
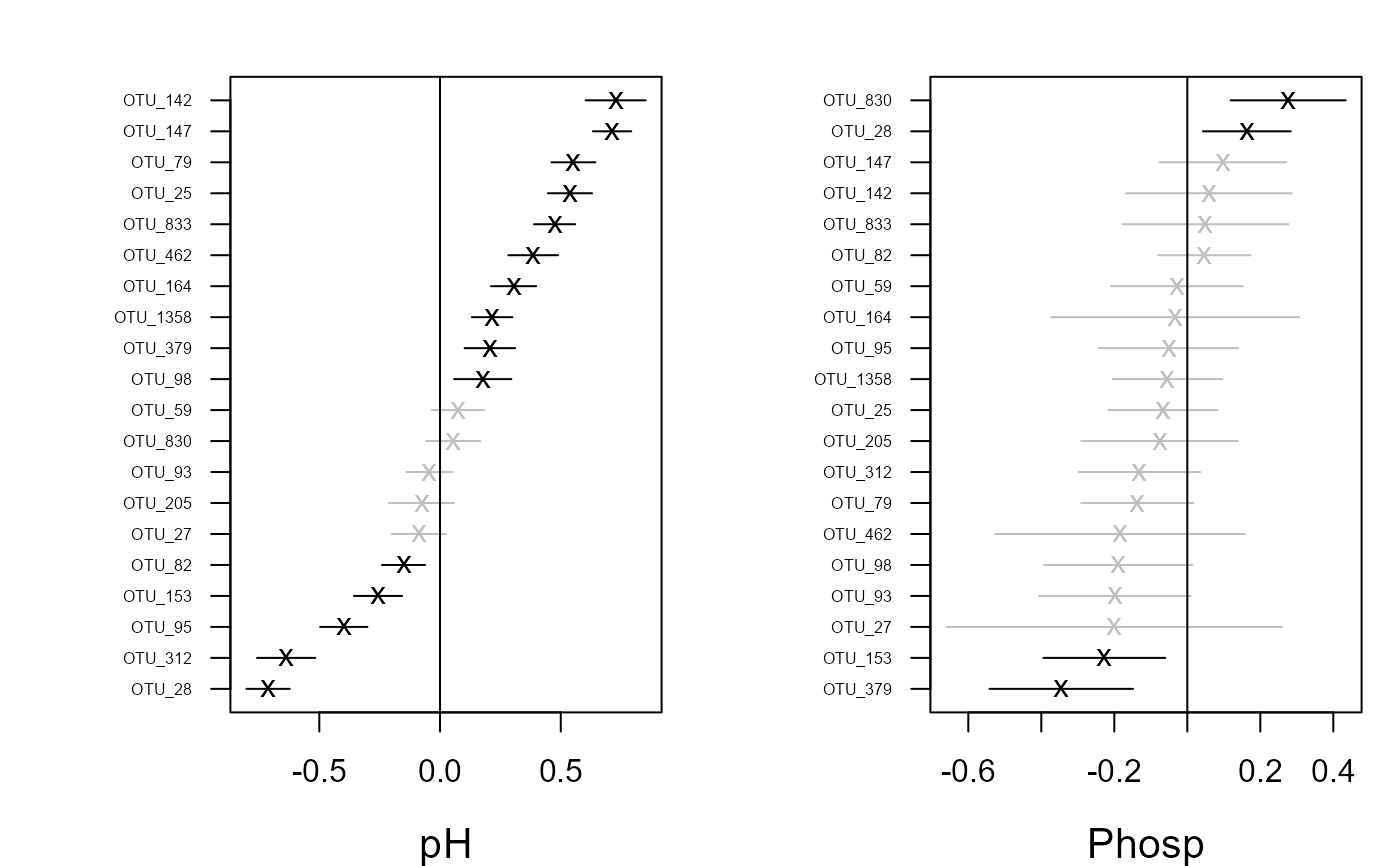
fit <- gllvm(y, X, formula = ~ pH + Phosp, family = poisson())
coefplot(fit)
 if (FALSE) { # \dontrun{
# Fit gllvm model with environmental covariances and reduced rank
fitRR <- gllvm(y = y, X = X, num.RR = 2, family = "negative.binomial")
coefplot(fitRR, mfrow=c(2,3))
} # }
if (FALSE) { # \dontrun{
# Fit gllvm model with environmental covariances and reduced rank
fitRR <- gllvm(y = y, X = X, num.RR = 2, family = "negative.binomial")
coefplot(fitRR, mfrow=c(2,3))
} # }